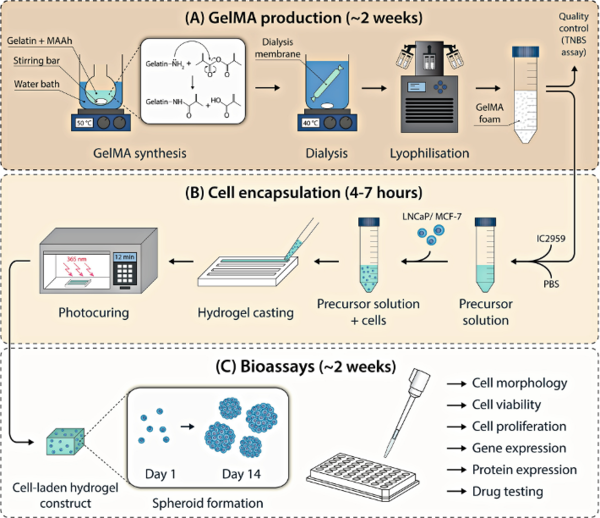
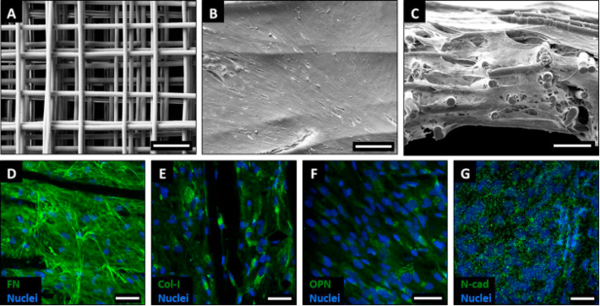
We use and fabricate tissue-engineered constructs that integrate extracellular matrix, molecular and biomechanical properties to grow functional tissues. For example, polycaprolactone (PCL) is used for melt electrospinning writing, a 3D printing technique, to generate scaffolds that allow the attachment of tissue-specific cell types. These PCL scaffolds provide a fibrous network and structural support for cell infiltration to mimic characteristic parameters of tissue-like features.
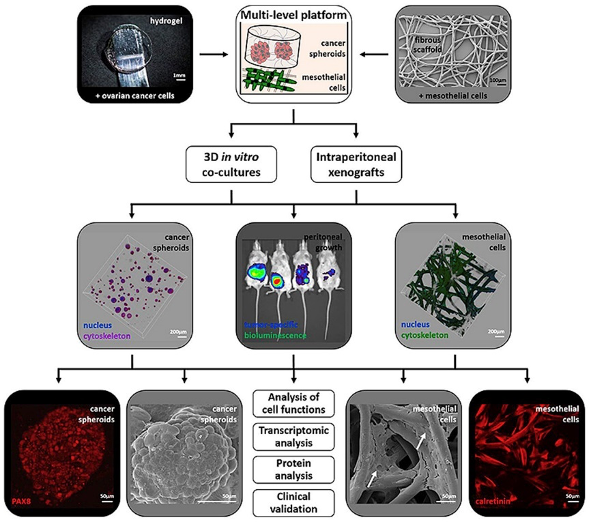
In an interdisciplinary setting, we design 3D approaches that deconstruct the native tissue composition and biomechanical properties of different tumour types to provide clinically predictive platforms and to test novel treatments. Therefore, we study the tumour biology and treatment responses of primary tumours, such as pancreatic cancer, ovarian cancer, colon cancer, neuroblastoma and osteosarcoma, as well as metastases, including peritoneal, prostate and breast cancer-induced metastasis.
SELECTED PUBLICATIONS (since 2020)
- Clegg, J., Curvello, R., Gabrielyan, A., Croagh, D., Hauser, S., Loessner, D. (2025) Tailoring metabolic activity assays for tumour-engineered 3D models. Biomaterials Advances 167, 214116, doi: 10.1016/j.bioadv.2024.214116.
- Curvello, R., Berndt, N., Hauser, S., Loessner, D. (2024) Recreating metabolic interactions of the tumour microenvironment. Trends in Endocrinology & Metabolism. doi: 10.1016/j.tem.2023.12.005.
- Curvello, R., Kast, V., Ordóñez-Morán, P., Mata, A., Loessner, D. (2023) Biomaterial-based platforms for tumour tissue engineering. Nature Reviews Materials 8(5), 314-330, doi: 10.1038/s41578-023-00535-3.
- Kast, V., Nadernezhad, A., Pette, D., Gabrielyan, A., Fusenig, M., Honselmann, K. C., Stange, D.E., Werner, C., Loessner, D. (2023) A tumor microenvironment model of pancreatic cancer to elucidate responses toward immunotherapy. Advanced Healthcare Materials 12(14), 2201907, doi: 10.1002/adhm.202201907.
- Osuna de la Peña, D., Trabulo, S. M. D., Collin, E., Liu, Y., Sharma, S., Tatari, M., Behrens, D., Erkan, M., Lawlor, R. T., Scarpa, A., Heeschen, C., Mata, A., Loessner, D. (2021) Bioengineered 3D models of human pancreatic cancer recapitulate in vivo tumour biology. Nature Communications 12(1), 5623, doi: 10.1038/s41467-021-25921-9.